EDA.ipynb
This notebook covers the exploratory data analysis of the tutorial which helps to gain necessary insights needed for selecting the best data preparation techniques, in order to get the best prediction results out of the machine learning model(s).
# importing required libraries and packages
import numpy as np
import pandas as pd
!pip3 install petroeval
import petroeval as pet
import matplotlib.pyplot as pltRequirement already satisfied: petroeval in c:\programdata\anaconda3\lib\site-packages (1.1.5.4)
Requirement already satisfied: matplotlib in c:\programdata\anaconda3\lib\site-packages (from petroeval) (3.3.2)
Requirement already satisfied: pandas in c:\programdata\anaconda3\lib\site-packages (from petroeval) (1.2.0)
Requirement already satisfied: numpy in c:\programdata\anaconda3\lib\site-packages (from petroeval) (1.19.2)
Requirement already satisfied: lasio==0.28 in c:\programdata\anaconda3\lib\site-packages (from petroeval) (0.28)
Requirement already satisfied: lightgbm in c:\programdata\anaconda3\lib\site-packages (from petroeval) (3.1.1)
Requirement already satisfied: xgboost in c:\programdata\anaconda3\lib\site-packages (from petroeval) (1.3.1)
Requirement already satisfied: certifi>=2020.06.20 in c:\programdata\anaconda3\lib\site-packages (from matplotlib->petroeval) (2020.6.20)
Requirement already satisfied: python-dateutil>=2.1 in c:\programdata\anaconda3\lib\site-packages (from matplotlib->petroeval) (2.8.1)
Requirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.3 in c:\programdata\anaconda3\lib\site-packages (from matplotlib->petroeval) (2.4.7)
Requirement already satisfied: cycler>=0.10 in c:\programdata\anaconda3\lib\site-packages (from matplotlib->petroeval) (0.10.0)
Requirement already satisfied: pillow>=6.2.0 in c:\programdata\anaconda3\lib\site-packages (from matplotlib->petroeval) (8.0.1)
Requirement already satisfied: kiwisolver>=1.0.1 in c:\programdata\anaconda3\lib\site-packages (from matplotlib->petroeval) (1.3.0)
Requirement already satisfied: pytz>=2017.3 in c:\programdata\anaconda3\lib\site-packages (from pandas->petroeval) (2020.1)
Requirement already satisfied: wheel in c:\programdata\anaconda3\lib\site-packages (from lightgbm->petroeval) (0.35.1)
Requirement already satisfied: scikit-learn!=0.22.0 in c:\programdata\anaconda3\lib\site-packages (from lightgbm->petroeval) (0.23.2)
Requirement already satisfied: scipy in c:\programdata\anaconda3\lib\site-packages (from lightgbm->petroeval) (1.5.2)
Requirement already satisfied: six>=1.5 in c:\programdata\anaconda3\lib\site-packages (from python-dateutil>=2.1->matplotlib->petroeval) (1.15.0)
Requirement already satisfied: joblib>=0.11 in c:\programdata\anaconda3\lib\site-packages (from scikit-learn!=0.22.0->lightgbm->petroeval) (0.17.0)
Requirement already satisfied: threadpoolctl>=2.0.0 in c:\programdata\anaconda3\lib\site-packages (from scikit-learn!=0.22.0->lightgbm->petroeval) (2.1.0)
Getting Data¶
The train data set comprise well logs from a total of 98 wells from the North Sea, while the open test data is made up of 10 wells, Only the gamma ray (GR) log is present in all the wells.
traindata = pd.read_csv('./data/train.csv', sep=';')
testdata = pd.read_csv('./data/leaderboard_test_features.csv.txt', sep=';')Data Exploration and Visualization¶
Here, the data is investigated to understand it better (shape and form). Since the data is a combination of 98 different wells, it will be time intensive to make visualizations of each of the wells, so a more combined approach into looking at the wells is taken. Both train and test data are explored simultaneously.
- Basic information check on wells and data
- Visualizing the percentage of each logs present in all wells
- Visualizing the percentage OF missing values of each logs present in all wells
- Spatial distribution of wells according to their coordinates
- Visualizing some log plots
traindata.head()traindata.shape, testdata.shape((1170511, 29), (136786, 27))With over 1million training data points, safe to call this "big data". Also, could be seen that the test data set has two logs lesser than the train data. Let's investigate that.
number = 0
logs = []
for log in traindata.columns:
if log not in testdata.columns:
logs.append(log)
number += 1
print(f'{number} logs not in test data:')
print(logs)2 logs not in test data:
['FORCE_2020_LITHOFACIES_LITHOLOGY', 'FORCE_2020_LITHOFACIES_CONFIDENCE']
traindata.columnsIndex(['WELL', 'DEPTH_MD', 'X_LOC', 'Y_LOC', 'Z_LOC', 'GROUP', 'FORMATION',
'CALI', 'RSHA', 'RMED', 'RDEP', 'RHOB', 'GR', 'SGR', 'NPHI', 'PEF',
'DTC', 'SP', 'BS', 'ROP', 'DTS', 'DCAL', 'DRHO', 'MUDWEIGHT', 'RMIC',
'ROPA', 'RXO', 'FORCE_2020_LITHOFACIES_LITHOLOGY',
'FORCE_2020_LITHOFACIES_CONFIDENCE'],
dtype='object')traindata.dtypesWELL object
DEPTH_MD float64
X_LOC float64
Y_LOC float64
Z_LOC float64
GROUP object
FORMATION object
CALI float64
RSHA float64
RMED float64
RDEP float64
RHOB float64
GR float64
SGR float64
NPHI float64
PEF float64
DTC float64
SP float64
BS float64
ROP float64
DTS float64
DCAL float64
DRHO float64
MUDWEIGHT float64
RMIC float64
ROPA float64
RXO float64
FORCE_2020_LITHOFACIES_LITHOLOGY int64
FORCE_2020_LITHOFACIES_CONFIDENCE float64
dtype: objectFrom above data types, there are three categorical logs denoted as 'object'. Getting more info on these logs will provide better insight to choose a better encoding technique.
print(f'Number of wells in data: {len(np.unique(traindata.WELL))}')Number of wells in data: 98
# checking more info on the categorical logs (FORMATION AND GROUP)
print(f'Unique formation count: {len(dict(traindata.FORMATION.value_counts()))}')
print(f'Unique group count: {len(dict(traindata.GROUP.value_counts()))}')Unique formation count: 69
Unique group count: 14
traindata.GROUP.value_counts()HORDALAND GP. 293155
SHETLAND GP. 234028
VIKING GP. 131999
ROGALAND GP. 131944
DUNLIN GP. 119085
NORDLAND GP. 111490
CROMER KNOLL GP. 52320
BAAT GP. 35823
VESTLAND GP. 26116
HEGRE GP. 13913
ZECHSTEIN GP. 12238
BOKNFJORD GP. 3125
ROTLIEGENDES GP. 2792
TYNE GP. 1205
Name: GROUP, dtype: int64traindata.FORMATION.value_counts()Utsira Fm. 172636
Kyrre Fm. 94328
Lista Fm. 71080
Heather Fm. 65041
Skade Fm. 45983
...
Broom Fm. 235
Intra Balder Fm. Sst. 177
Farsund Fm. 171
Flekkefjord Fm. 118
Egersund Fm. 105
Name: FORMATION, Length: 69, dtype: int64Checking for the percentage of each logs present in all wells¶
For train data
# this code is extracted from Matteo Niccoli github EDA repo for the FORCE competition:
# https://github.com/mycarta/Force-2020-Machine-Learning-competition_predict-lithology-EDA/blob/master/Interactive_data_inspection_and_visualization_by_well.ipynb
occurences = np.zeros(25)
for well in traindata['WELL'].unique():
occurences += traindata[traindata['WELL'] == well].isna().all().astype(int).values[2:-2]
fig, ax = plt.subplots(1, 1, figsize=(14, 7))
ax.bar(x=np.arange(occurences.shape[0]), height=(traindata.WELL.unique().shape[0]-occurences)/traindata.WELL.unique().shape[0]*100.0)
ax.set_xticklabels(traindata.columns[2:-2], rotation=45)
ax.set_xticks(np.arange(occurences.shape[0]))
ax.set_ylabel('Well presence (\%)')Text(0, 0.5, 'Well presence (\\%)')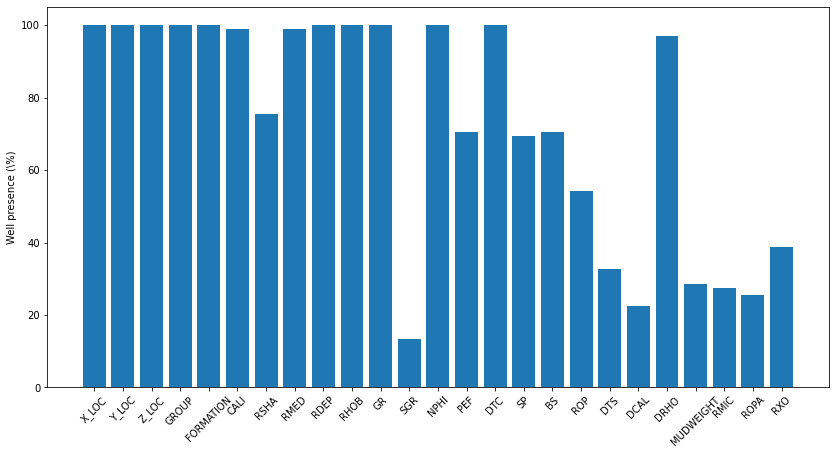
# this code is extracted from Matteo Niccoli github EDA repo for the FORCE competition:
# https://github.com/mycarta/Force-2020-Machine-Learning-competition_predict-lithology-EDA/blob/master/Interactive_data_inspection_and_visualization_by_well.ipynb
occurences = np.zeros(23)
for well in testdata['WELL'].unique():
occurences += testdata[testdata['WELL'] == well].isna().all().astype(int).values[2:-2]
fig, ax = plt.subplots(1, 1, figsize=(14, 7))
ax.bar(x=np.arange(occurences.shape[0]), height=(testdata.WELL.unique().shape[0]-occurences)/testdata.WELL.unique().shape[0]*100.0)
ax.set_xticklabels(testdata.columns[2:-2], rotation=45)
ax.set_xticks(np.arange(occurences.shape[0]))
ax.set_ylabel('Well presence (\%)')Text(0, 0.5, 'Well presence (\\%)')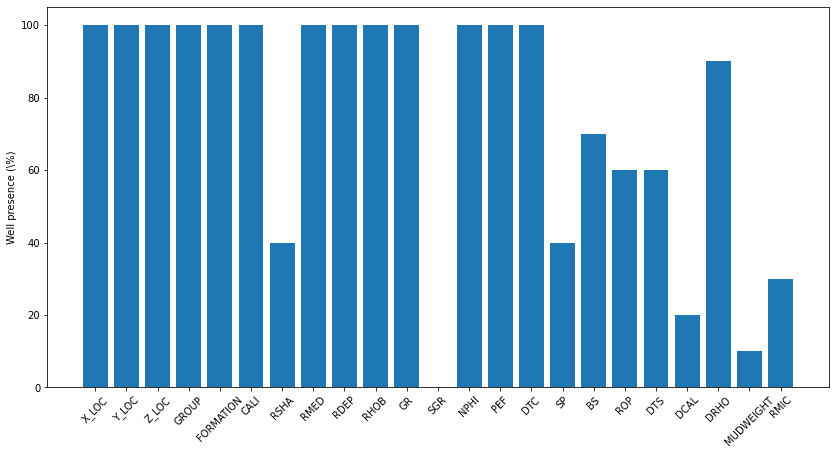
Logs are presently different in both train and test data, as we can see, the SGR log is absent in all test logs
While figure above shows the log presence based on appearance on all wells, the below shows the percentage of logs based on missing values/actual data present
train_well_items = dict(100 - (traindata.isna().sum()/traindata.shape[0]) * 100)
test_well_items = dict(100 - (testdata.isna().sum()/testdata.shape[0]) * 100)train_well_items = {log:value for log, value in train_well_items.items() if value != 100.0}
test_well_items = {log:value for log, value in test_well_items.items() if value != 100.0}train_well_items{'X_LOC': 99.07946187605242,
'Y_LOC': 99.07946187605242,
'Z_LOC': 99.07946187605242,
'GROUP': 99.89081691671416,
'FORMATION': 88.29622276082839,
'CALI': 92.49242424889643,
'RSHA': 53.878177992346934,
'RMED': 96.66871990096632,
'RDEP': 99.05895801064663,
'RHOB': 86.22234220780497,
'SGR': 5.925019072866462,
'NPHI': 65.3910129849271,
'PEF': 57.38450984228256,
'DTC': 93.09164971538073,
'SP': 73.83501735566773,
'BS': 58.32128019300972,
'ROP': 45.712599027262456,
'DTS': 14.91767270875711,
'DCAL': 25.530131711705394,
'DRHO': 84.3953623673763,
'MUDWEIGHT': 27.0096564662784,
'RMIC': 15.049837207851951,
'ROPA': 16.430857975704626,
'RXO': 27.972996409260574,
'FORCE_2020_LITHOFACIES_CONFIDENCE': 99.98470753371818}occurences = np.zeros(len(train_well_items))
fig, ax = plt.subplots(1, 1, figsize=(14, 7))
ax.bar(x=np.arange(occurences.shape[0]), height=train_well_items.values())
ax.set_xticklabels(train_well_items.keys(), rotation=45)
ax.set_xticks(np.arange(occurences.shape[0]))
ax.set_ylabel('Data presence (\%)')Text(0, 0.5, 'Data presence (\\%)')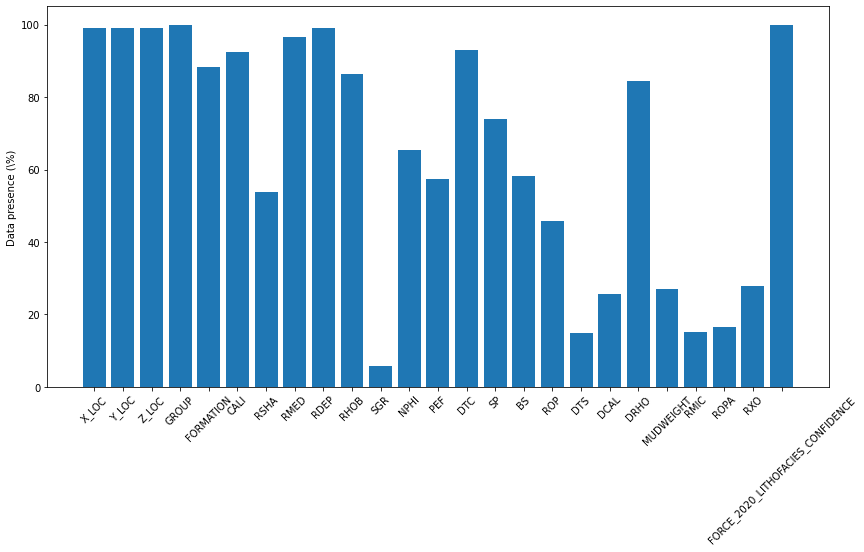
occurences = np.zeros(len(test_well_items))
fig, ax = plt.subplots(1, 1, figsize=(14, 7))
ax.bar(x=np.arange(occurences.shape[0]), height=test_well_items.values())
ax.set_xticklabels(test_well_items.keys(), rotation=45)
ax.set_xticks(np.arange(occurences.shape[0]))
ax.set_ylabel('Data presence (\%)')Text(0, 0.5, 'Data presence (\\%)')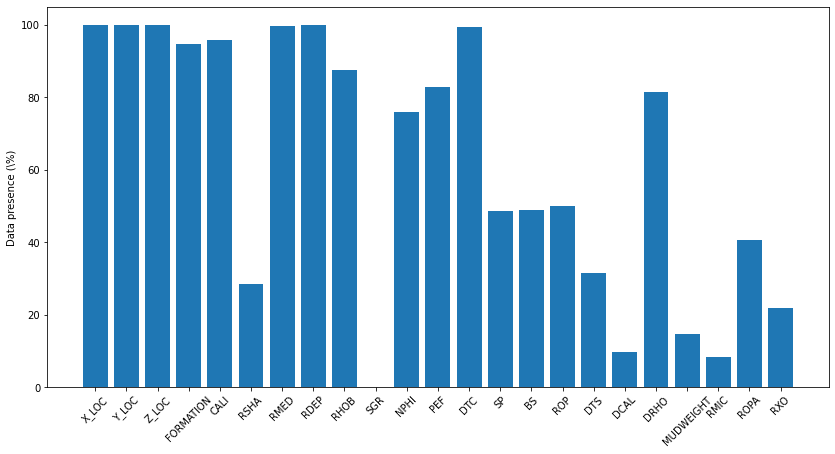
Visualizing the train labels categories
labels = dict(traindata.FORCE_2020_LITHOFACIES_LITHOLOGY.value_counts())lithofacies_names = ['Shale', 'Sandstone', 'SS/Shale', 'Marl',
'Dolomite', 'Limestone', 'Chalk', 'Halite', 'Anhydrite',
'Tuff', 'Coal', 'Basement']traindata.FORCE_2020_LITHOFACIES_LITHOLOGY.value_counts()65000 720803
30000 168937
65030 150455
70000 56320
80000 33329
99000 15245
70032 10513
88000 8213
90000 3820
74000 1688
86000 1085
93000 103
Name: FORCE_2020_LITHOFACIES_LITHOLOGY, dtype: int64fig = plt.figure(figsize=(15, 10))
plt.bar(lithofacies_names, (np.array(list(labels.values()))/traindata.shape[0]) * 100)<BarContainer object of 12 artists>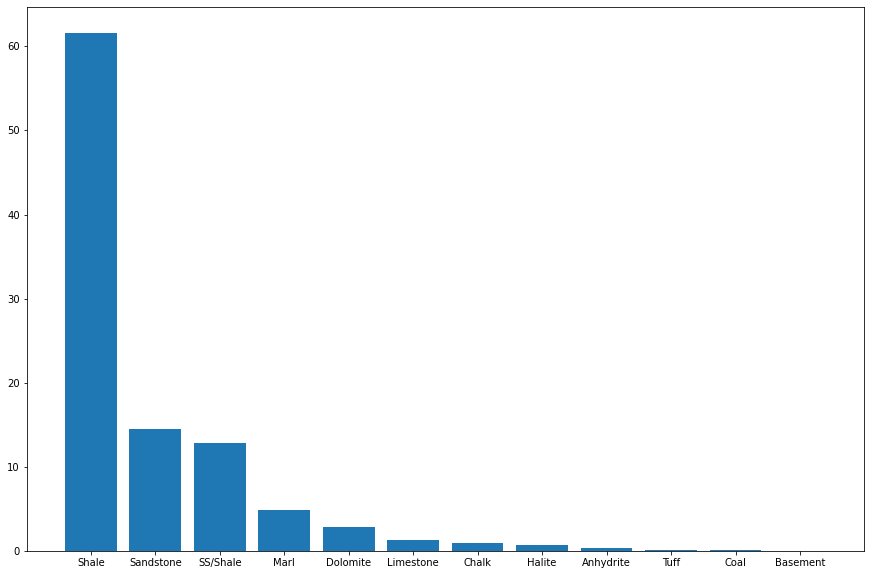
# spatial distribution of both train and test wells
fig = plt.figure(figsize=(12,8))
plt.scatter(traindata.X_LOC, traindata.Y_LOC, c='g')
plt.scatter(testdata.X_LOC, testdata.Y_LOC, c='r')
plt.show()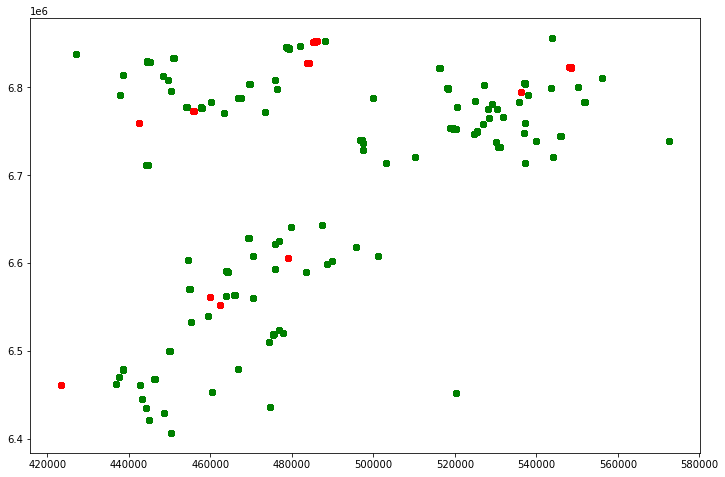
From the plot, we can see that the test wells are evenly distributed to cover the train wells spread. This makes it easier while preparing the data sets as we get to apply almost same techniques to both data sets.
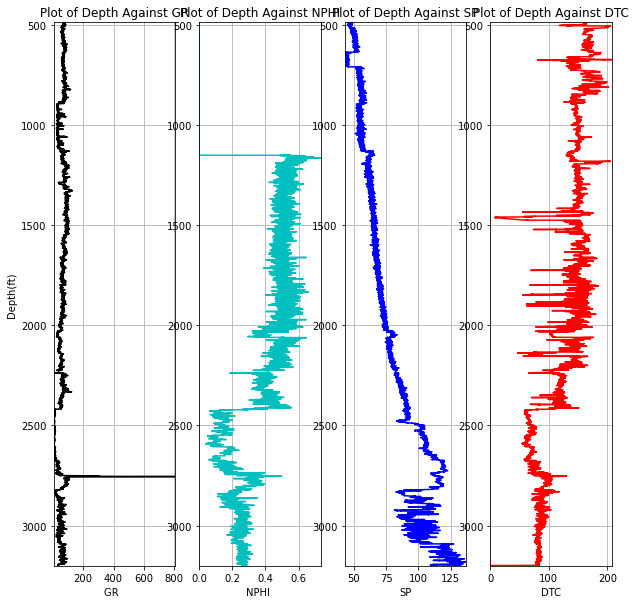
While it might be impossible to view all train and test logs, let's take a look at a test well in close proximity to the train logs and compare log signatures
testdata.loc[testdata.WELL == '15/9-14']pet.four_plots(testdata.loc[testdata.WELL == '15/9-14'], x1='GR', x2='NPHI', x3='SP', x4='DTC',
top=480, base=3565, depth='DEPTH_MD')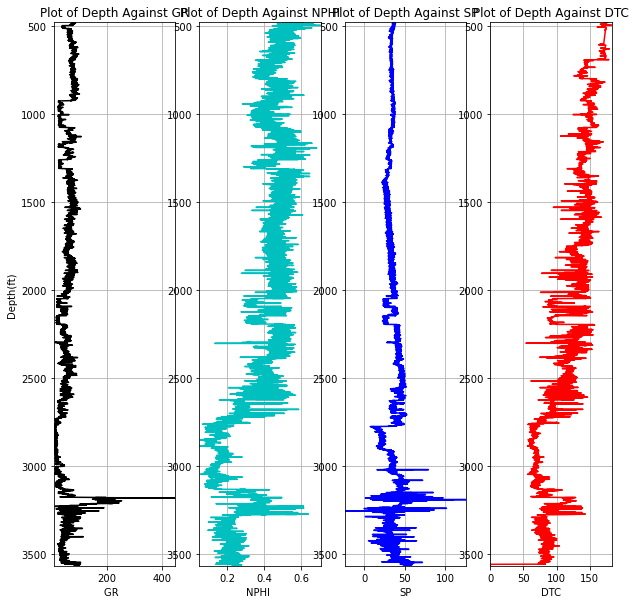
traindata.WELL.unique()array(['15/9-13', '15/9-15', '15/9-17', '16/1-2', '16/1-6 A', '16/10-1',
'16/10-2', '16/10-3', '16/10-5', '16/11-1 ST3', '16/2-11 A',
'16/2-16', '16/2-6', '16/4-1', '16/5-3', '16/7-4', '16/7-5',
'16/8-1', '17/11-1', '25/11-15', '25/11-19 S', '25/11-5',
'25/2-13 T4', '25/2-14', '25/2-7', '25/3-1', '25/4-5', '25/5-1',
'25/5-4', '25/6-1', '25/6-2', '25/6-3', '25/7-2', '25/8-5 S',
'25/8-7', '25/9-1', '26/4-1', '29/6-1', '30/3-3', '30/3-5 S',
'30/6-5', '31/2-1', '31/2-19 S', '31/2-7', '31/2-8', '31/2-9',
'31/3-1', '31/3-2', '31/3-3', '31/3-4', '31/4-10', '31/4-5',
'31/5-4 S', '31/6-5', '31/6-8', '32/2-1', '33/5-2', '33/6-3 S',
'33/9-1', '33/9-17', '34/10-19', '34/10-21', '34/10-33',
'34/10-35', '34/11-1', '34/11-2 S', '34/12-1', '34/2-4',
'34/3-1 A', '34/4-10 R', '34/5-1 A', '34/5-1 S', '34/7-13',
'34/7-20', '34/7-21', '34/8-1', '34/8-3', '34/8-7 R', '35/11-1',
'35/11-10', '35/11-11', '35/11-12', '35/11-13', '35/11-15 S',
'35/11-6', '35/11-7', '35/12-1', '35/3-7 S', '35/4-1', '35/8-4',
'35/8-6 S', '35/9-10 S', '35/9-2', '35/9-5', '35/9-6 S', '36/7-3',
'7/1-1', '7/1-2 S'], dtype=object)traindata.loc[traindata.WELL == '15/9-15']pet.four_plots(traindata.loc[traindata.WELL == '15/9-15'], x1='GR', x2='NPHI', x3='SP', x4='DTC',
top=485, base=3201, depth='DEPTH_MD')